How to Use
1. Search and View
There are 4 ways to search and view the SNP data in Human Genetic Variation Browser. The way to search from top page is explained below.1-1. Keyword Search
(1)Enter keywords at search box "Gene name / ID" and press GO button.(2)Search result is shown as below. The viewer is shown by clicking the link in the column of Gene Symbol. The link in the column of Gene ID will lead you to Gene in NCBI. The details of viewer is described in section 2. Viewer.
1-2. Search by rsID
(1)Enter the rsID of dbSNP in the box of dbSNP rsID and press GO button.(2)The search result is shown as below. The viewer opens by clicking the View link. The page of NCBI dbSNP opens by clicking the link in the column of rsID. The details of viewer is described in section 2. Viewer.
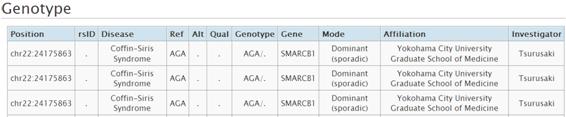
1-3. Search by Pathogenic Variation
(1)Enter keywords in the box of Pathogenic Variation and press GO button. You can search the pathogenic variation information registered in this database system. Candidate list of genes and diseases is shown when you enter a keyword.(2)Search result is shown as below. Viewer is shown by clicking the link.
1-4. Search by Chromosome
Clicking the number of chromosome leads you directly to the viewer.2. Viewer
The viewer shows the graphical image of SNP data. It contains the information of gene, dbSNP, exome, allele frequency, eQTL and so on.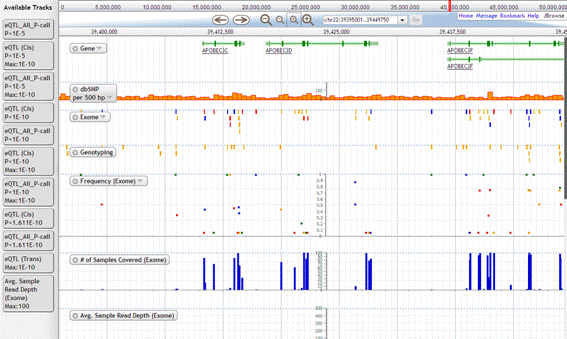
2-1. Operation Method
Move
There are 2 ways to move the viewer.
1. Drag the viewer and move right or left.
2. Press the arrow button at the top of the viewer.

Scaling
There are 2 ways to scale the viewer.
1. Press the magnifier button at the top of the viewer.

2. Drag the position bar and select the region to scale up. After releasing the mouse button, the viewer zooms up.
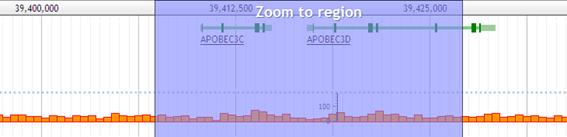
Move to other region on the same chromosome
Drag the bar of chromosome region to scale up.After releasing the mouse button, the viewer zooms up.

Display or hide the track
There are 2 ways to display or hide the track in the viewer.
1. The track is hidden by clicking the X button at the label.
2. Drag the items in the Available Tracks and drop it at the viewer, the track is shown.
2-2. Track
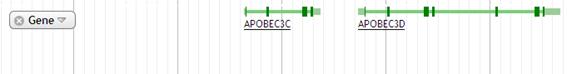
1. Gene
Gene track shows the gene information. The page of NCBI Gene is shown by clicking this track.
2. dbSNP
This track shows the NCBI dbSNP data. SNP is shown in yellow. Ins/del is shown in pink. The page of NCBI dbSNP is shown by clicking this track.

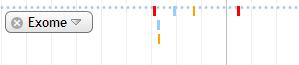
3. Exome
Exome which matches with dbSNP is shown in yellow. Green indicates the exome that matches with chip. Blue is the exome that matches both dbSNP and chip. Unique exome is shown in red.
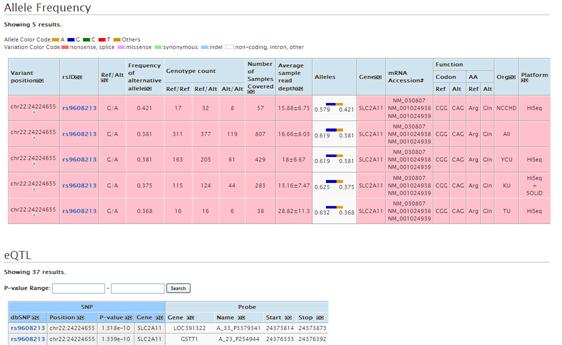
Allele frequency and eQTL data are shown by clicking the viewer. Allele frequency is collor-coded according to the type of variation such as nonsense and missense.
4. Genotyping
This track shows the genotyping data.

Detail information is shown by clicking.

5. Frequency(Exome)
Allele frequency of exome is shown in this track. The plot is shown in different color by base. The frequency of variant allele is plotted at the some level of scale. The frequency of both variant and reference allele is shown in pie chart otherwise.
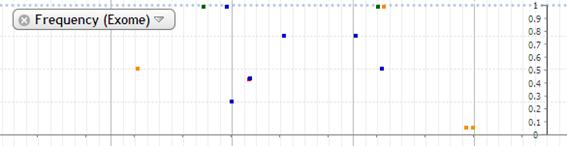
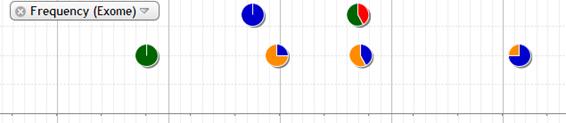
The detail data is popped up by mouse-over.
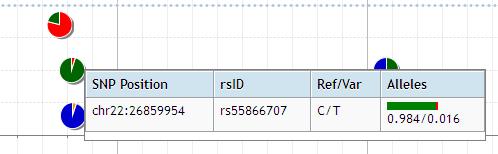
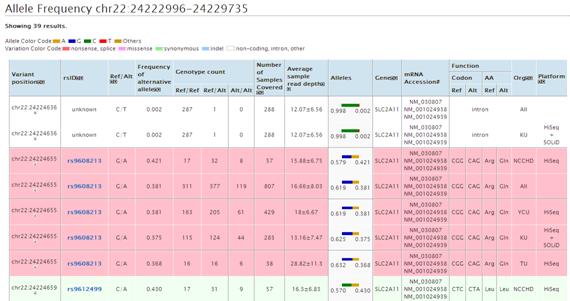
At the some zoom level, the more infomation is shown by clicking the plot as below. Double clicking the plot opens the new page to show the all frequency information in the area displayed in viewer. The data is color-coded according to the type of variation.
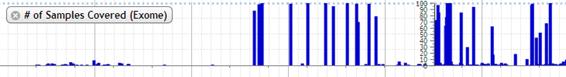
6. # of Samples Covered (Exome)
The number of samples which is both called and not called is shown in bar chart.
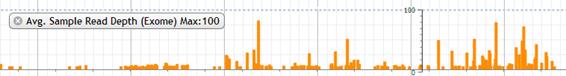
7. Avg. Sample Read Depth (Exome)
The average of sample read depth which is both called and not called is shown in bar chart.
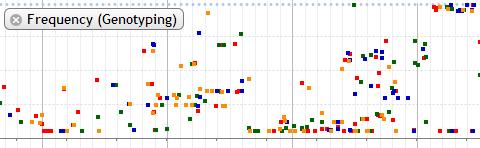
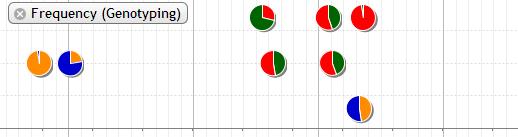
8. Frequency (Genotyping)
The genotyping data is shown in dot plot and pie chart.
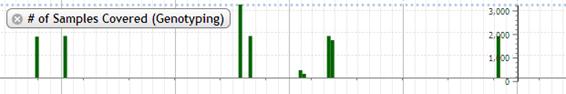
9. # of Samples Covered (Genotyping)
The genotyping data which is called is shown in bar chart.
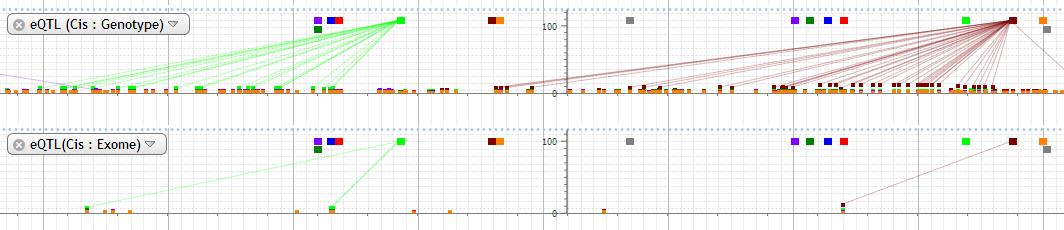
10. eQTL (Cis: Genotype), eQTL(Cis: Exome)
eQTL data based on genotype and exome data are available. The log of eQTL p-value which is multiplied by -1 is plotted. The probe is indicated by square in different colors at the top of plot. The color of probe and every dot corresponds to each other. Probe and dot which is under 10-5 is connected with line by default.
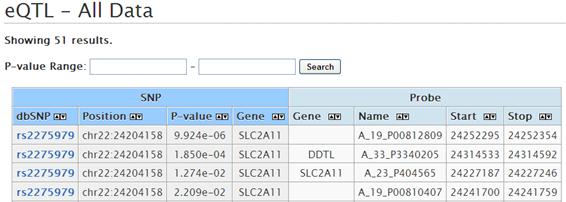
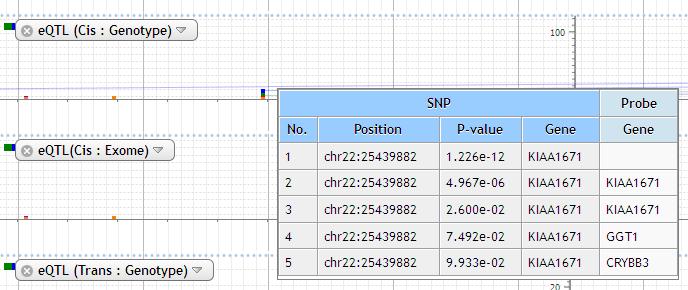
At the some zoom level, more information is displayed by clicking the plot and mouse-over.
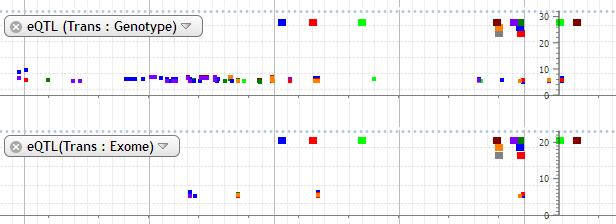
11. eQTL (Trans: Genotype), eQTL(Trans: Exome)
eQTL data based on genotype and exome data are available. The log of eQTL p-value which is multiplied by -1 is plotted. The data with P value less than 10 -5 is shown on the plot. The probe is indicated by square in different colors at the top of plot. The color of probe and every dot corresponds to each other. Probe and dot is not connected with line.
12. Pathogenic variation
The pathogenic variation data is shown in this track. The registered user can register the data of pathogenic variation.